Definition of Restriction Endonucleases
- Restriction endonucleases are enzymes that cut DNA at specific sequences, acting as molecular scissors.
- These enzymes are naturally found in bacteria, where they protect against foreign DNA (e.g., viral DNA) by cutting it.
Types of Restriction Enzymes
-
Type I
- Cut DNA at random locations, often hundreds of base pairs away from their recognition site.
- Less useful for precise genetic engineering.
-
Type II
- The most commonly used type in laboratories.
- Cut within or at short distances from their recognition sites, producing predictable and reproducible fragments.
- Recognition sites are often palindromic (the same sequence read 5′→3′ on both strands).
-
Type III
- Cut DNA at sites a short distance away (about 20–30 base pairs) from the recognition site.
- Less frequently used in routine cloning compared to Type II.
Common Restriction Enzymes and Their Recognition Sites
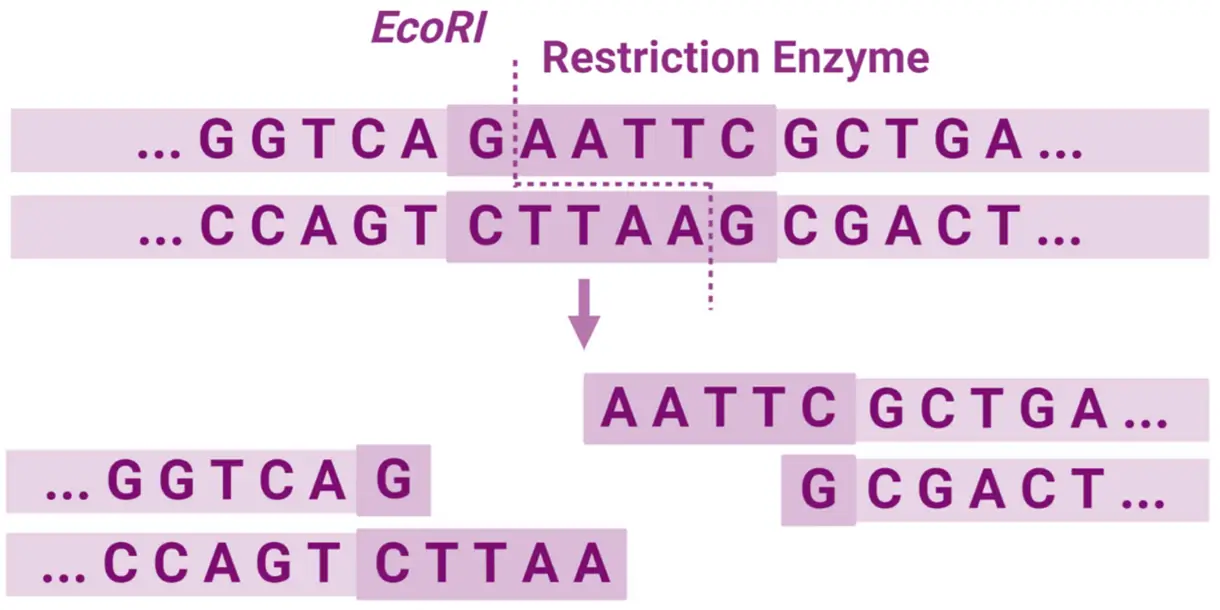
- EcoRI → Recognizes 5′-GAATTC-3′ and produces sticky ends.
- HindIII → Recognizes 5′-AAGCTT-3′ and produces sticky ends.
- HaeIII → Recognizes 5′-GGCC-3′ and produces blunt ends.
Types of DNA Ends After Cutting
-
Sticky Ends (Cohesive Ends)
- The enzyme cuts the two DNA strands at offset positions, leaving overhanging single-stranded regions.
- These overhangs can base-pair (anneal) with complementary overhangs cut by the same enzyme.
-
Blunt Ends
- The enzyme cuts both DNA strands at the same position, leaving no single-stranded overhang.
- While this creates more stable DNA ends, blunt-end ligations can be less efficient compared to sticky-end ligations.
- Sticky Ends → Single-stranded overhangs that help in DNA ligation.
- Blunt Ends → No overhangs, making ligation less efficient.
Click Here to Watch the Best Pharma Videos